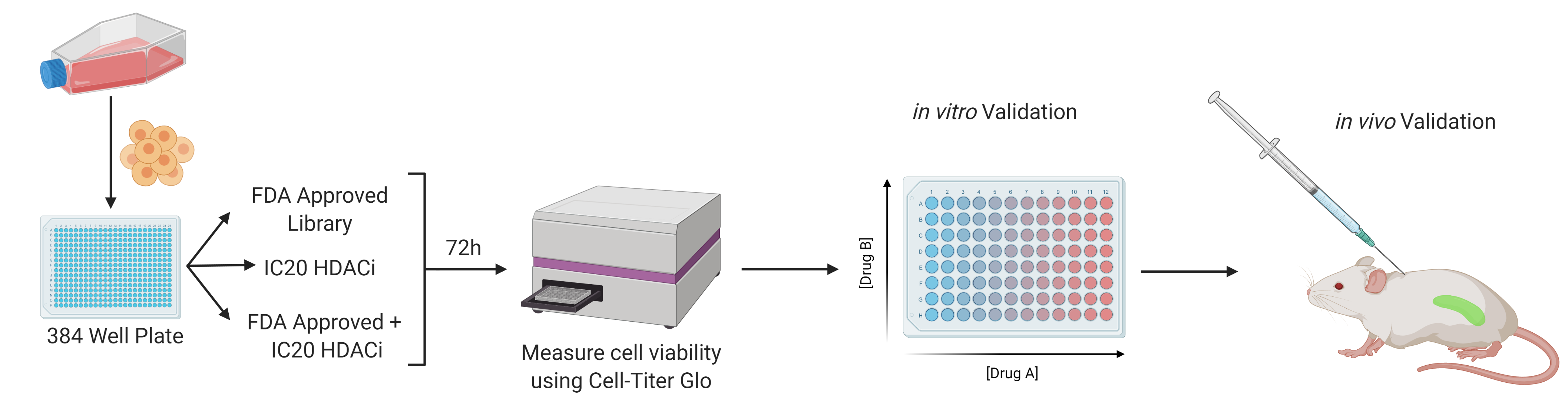
T-Cell Lymphomas (TCL) are a heterogeneous group of highly aggressive mature non-Hodgkin lymphomas that has its genesis from the malignant transformation of mature T-cells. Unfortunately, TCLs are generally associated with poor prognosis and most of the current therapies show limited efficacy and none are curative, making relapsed/resistant TCL a major clinical problem. One promising line of therapy currently being explored in the field involves the use of an epigenetic class of therapeutics, histone deacetylase inhibitors (HDACi). However, many patients will not respond to HDACi treatment and among those that do, resistance is inevitable. Using a well-defined bank of T-cell lymphoma patient samples undergoing HDACi therapy, I aim to identify the underlying genetic aberrations that lead to HDACi resistance. Through these results, I hope to improve out understanding of TCL pathogenesis during HDACi treatment and to develop novel strategies to overcome resistance.